Jon Massey - Blog
Jon Massey’s Personal Blog
Ontological Subdomains of Veterinary Medicines
A large proportion of my PhD has been spent dealing with data regarding the sales, usage, licensing, and constitution of veterinary medicines. There appears to be a great diversity in the way in which veterinary medicines are identified in the different sources of data which I have acquired along the way - name of medicine, supplier product code, Global Trade Identification Number (GTIN, i.e. the product barcode), Veterinary Medicine Number (VMNo - more on this later). In trying to reconcile these differences and conversing with a variety of people who interact with veterinary medicines it has led me to the question:
What even is a veterinary medicine?
Bear with me, while I take you on a journey into the ontology of veterinary medicines and routes to greater clarity in this domain.
Identifying Veterinary Medicines
With regard to the identification numbers/codes outlined above, I have been seeking authoritative open mappings between these sets of identifiers and have so far come up short. Identifying a product by name presents a separate set of issues and is a blog post for another time on applications of Natural Language Processing (NLP) for Named Entity Recognition (NER), but the issues of relevance here are that of what reference set of entities one matches to and the issue of ambiguous matches/insufficient information for unambiguous matches. In all of my medicine identification efforts, I have been using the Veterinary Medicine Directorate’s Product Information Database (VMD PID) which is the official reference of veterinary medicines licensed for sale in the UK.
The Product Information Database and VM Numbers
The VMD PID contains information regarding all veterinary medicines currently licensed for sale in the UK, those with expired licenses, those with suspended licenses and homoeopathic preparations. The primary identifier of each record is the VMNo (see above), which is also referred to as the Marketing Authorisation Number (MANo) in literature such as Summary Product Characteristics (SPC) documents for products. In using this database as a reference set for my NLP and other works, I was finding that this number was changing over time for certain products, which was causing me issues in validating my NER techniques. This was a source of great frustration for me: why were the identifiers for a product changing when the product was staying the same…?
The answer, of course, was that the product records weren’t staying the same but were changing in ways that weren’t obvious or important to me. I wrote a small program to detect and check these changes, and found that over a period covering July to October 2019 there were 24 instances where a licensed veterinary medical product changed VMNo yet its name stayed the same. In many of these cases, an attribute relating to the marketing authorisation of the product, e.g. the name or address of the licensee, had changed. This revealed to me that the entities in the Product Information Database where not, in fact, veterinary medicines but Marketing Authorisations of veterinary medicines. This ontological mis-match (or impedance mismatch in technical jargon) was the source of the confusion and frustration.
Seeking clarity in the ontology of veterinary medicines
Posing the question at the head of this article to a number of people who use veterinary medicines or work with datasets regarding them gave a surprising range of answers. I’d like to present three subdomains with three different conceptions of a veterinary medicine (i.e. what are the defining characteristics of a veterinary medicine that you might use to tell one from another) into which most of these answers coalesced.
The functional domain
What is it, where do you stick it, and what does it do?
Actors in this domain are primarily concerned with the functional characteristics of veterinary medicines - what species can the product be used in, to treat what conditions, by which route of administration.
The analytical domain
What’s in it, and how much?
Actors in this domain are primary concerned with the quantitative composition of veterinary medicines - the physical preparation of the medicine, the active ingredients, excipients and their concentrations.
The regulatory domain
Who has the rights to market this product, and for what purposes?
Actors in this domain are primarily concerned with the conditions under which veterinary medicines are licensed for sale.
The differences between these conceptions of what a veterinary medicine is reveal themselves in these differences in identification schemes used in these domains. In the absence of a clear singular definition of what a veterinary medicine is, is there an ontology of the concepts around veterinary medicines which could allow for more precise definition and identification of the entities therein which transcends these subdomains?
Existing ontologies and next steps
ATCVet
The Anatomical Therapeutic Chemical classification system for veterinary medicinal products, ATCvet, is based on the same main principles as the ATC classification system for drug substances used in human medicine.
– The ATCVet Classification System, WHOCC
The ATCVet system is a hierarchical taxonomy of veterinary medicines produced by the WHO Collaborating Centre for Drug Statistics Methodology. As the name suggests, medicines are first categorised by the anatomical structure used as the route of administration (e.g. QD - Dermatologicals), then by the therapeutic indication (e.g. WD06 - Antibiotics and chemotherapeutics for dermatological use), then by increasingly precise descriptions of their chemical components (e.g. QD06AA - Tetracyline and Derivatives > QD06AA02 - Chlortetracyline). Such a classification is useful for those in the analytical domain and to a certain extent those in the functional domain but lacks information regarding specific clinical indications, physical formulations, quantitative compositional information, or any species indications. Equally, there is varying levels of precision in different branches of the terminal chemical layer - compare the above given precise examples with the imprecise definition of QJ51RV02 - “antibacterials, antimycotics and corticosteroids”. Nonetheless, the ATCvet system does go some way to providing a structured classification of veterinary medicines and is easily navigable.
RXNorm
RXNorm began as a project of the National Library of Medicine in 2005 as a standardised vocabulary of drug names in human medicine. It has subsequently been integrated into the Unified Medical Language System which provides a very large and comprehensive structured dataset of medical concepts. A good illustration of the concepts related to medicines is given in the below example as shown through the RXNav viewer.
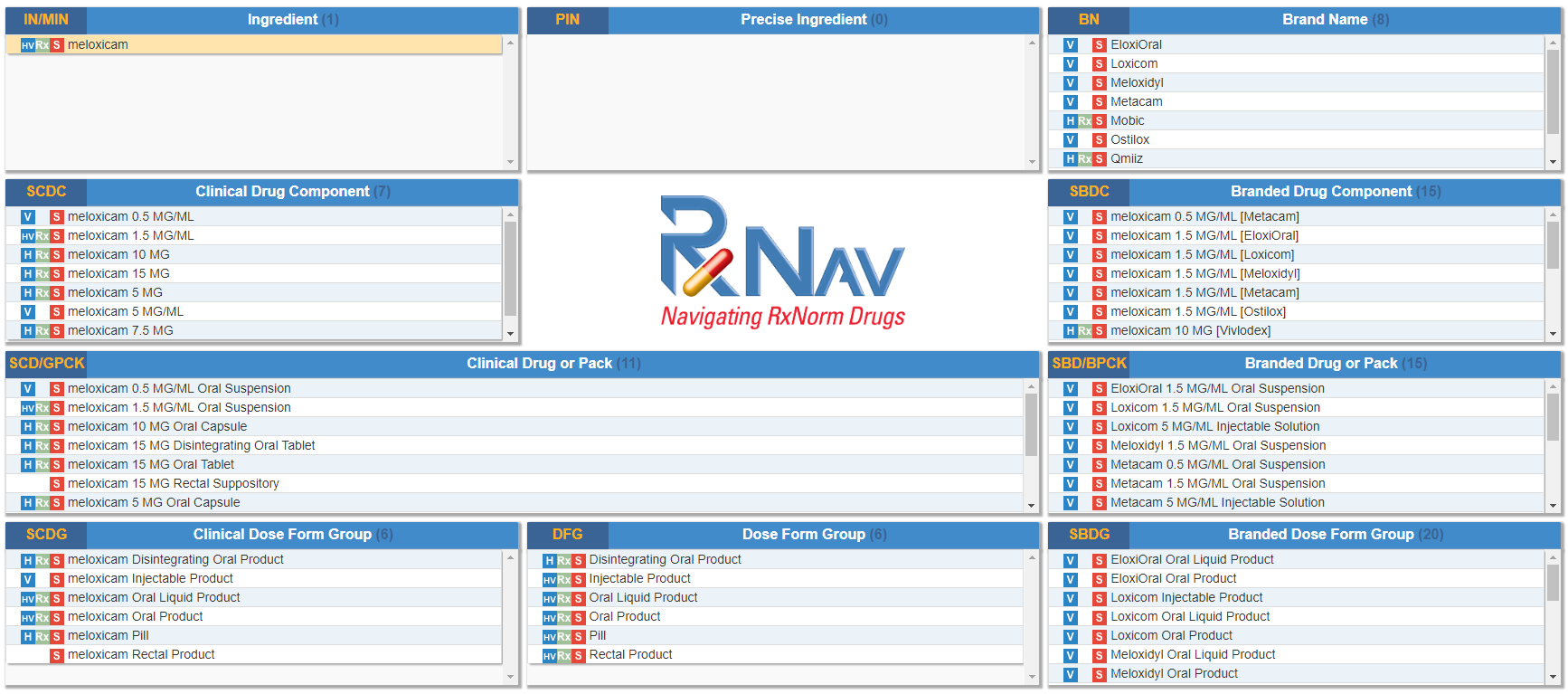
A full listing of the RXNorm term types is given in the RXNorm documentation, which is well-defined and comprehensive. Due to the fact that some veterinary data sources, such as VetSCT, some veterinary medicine data is currently present in RXNorm (e.g. Enrofloxacin). Unfortunately this provision is incomplete from a veterinary medicine concern, there are omissions of multi-species concepts (a.k.a “term types”) and terms within these concepts (e.g. intramammary dose forms are absent yet are common in veterinary medicine).
One of the fundamental differences between human and veterinary medicine is the fact that in human medicine only one species is treated whereas in veterinary medicine multiple species are treated. The implications of this on a medicine ontology is that certain products are only licensed for use in certain species. Unfortunately there are further dimensions to this species-medicine relationship, since certain medicines are only licensed for treatment of certain species via certain routes of administration and for certain indications. Additionally, in the UK at least, veterinary surgeons are permitted to prescribe veterinary medicines outside of the conditions of the license in what is known as the Cascade System. An example which illustrates this complexity is that of Linco-Spectin a combination aminoglycoside antibiotic licensed for oral use in pigs and poultry than is also used topically in cattle as a foot bath treatment. Lincomycin is toxic to ruminants if given orally, thus highlighting the importance of the species-specificity of the given indications and routes of administration.
In order to extend RXNorm for full coverage of veterinary concepts and terms, great consideration should be given to how best to structure these concepts and which information to include in what way. Common usage of a veterinary medicine for a particular indication under the cascade system is useful information to both veterinary and one health researchers, but should be clearly distinct from a specific license for an indication. Chemical ingredients within RXNorm are well specified with links to reference datasets such as PubChem, but a further extension that may be particularly useful to One Health AMR researchers would be linkage to a taxonomy of antimicrobial compounds. Such linkage would facilitate the answering of queries such as “for a given human medicine, which medicines of the same antimicrobial class are used in which animal species?”.
Such an extension project would be a useful resource to researchers, I believe. How best we might achieve it remains to be seen!